|
FAQs - Frequently asked questions
|
|
 Expand All Expand All  Collapse All Collapse All
|
| |
 1. What is Human-gpDB and what information does it contain? 1. What is Human-gpDB and what information does it contain? |
Human-gpDB is a publicly accessible, relational database of human G-Proteins and their interactions with human GPCRs and Effectors. The database currently holds information about 713 human GPCRs, 36 human G-Proteins and 99 human Effectors. Human-gpDB reveals information about 1663 connections between GPCRs and G-Proteins and 1618 connections between G-Proteins and Effectors. Advanced data integration techniques make Human-gpDB very rich in context since all of the bioentities are linked to a rich variety of external data sources. High quality visualization methods make the networks more informative and the extraction of information easier. Human-gpDB is currently a very useful tool for drug targeting investigation.
|
|
|
 2. How did we generate the data? 2. How did we generate the data? |
In order to collect information concerning the interactions between GPCRs, G-Proteins and Effectors in Human-gpDB, we performed an extensive literature search. The initial sequence information for all the molecules was retrieved from the UNIPROT database. The entries were acquired using suitable scripts written in Perl in order to parse the DE (description), the GN (gene) or the DR (database cross reference) field in the respective database entry. The datasets were then checked in order to eliminate duplicates. We used user-written Perl scripts to manipulate the data, whereas annotations regarding the interaction between G-proteins and effectors, the effect of the particular interaction and the corresponding references were appended manually in a spreadsheet. For all the interactions, we provide links to PUBMED corresponding to original articles reporting the association. |
|
|
 3. How is data structured? 3. How is data structured? |
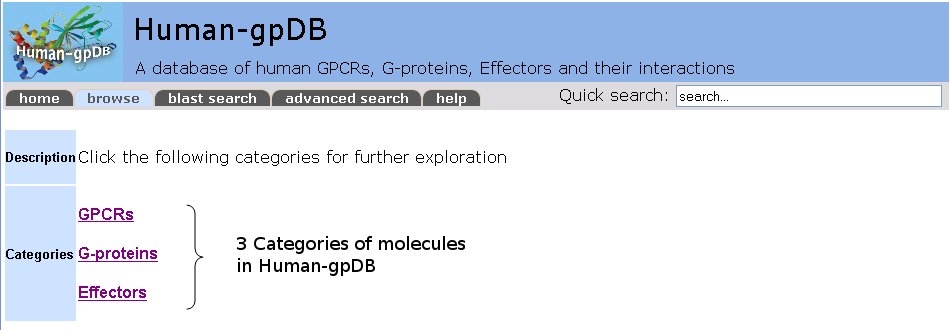
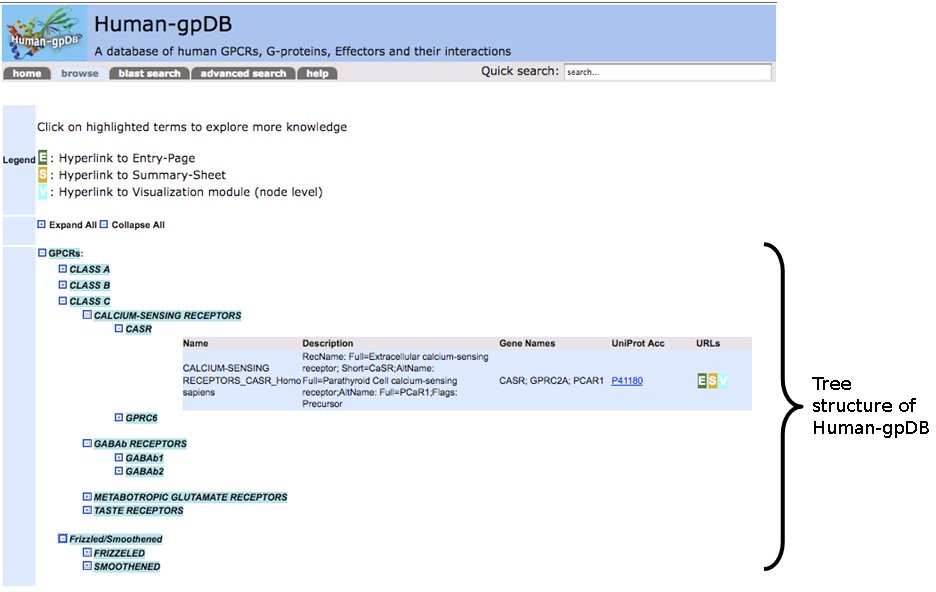
Human-gpDB is organized in a tree structure to separate data onto different layers. We use hierarchies to show the classification of the proteins. G-Proteins and GPCRs are classified according to a hierarchy of different classes, families and sub-families, whereas Effectors are classified in families, subfamilies and types, based on extensive literature search. The classification of GPCRs follows the IUPHAR classification, while the Effectors classification is a unique feature and is based on their function. The user can expand or collapse the tree at will.
 |
|
|
 4. How do we use Human-gpDB? 4. How do we use Human-gpDB? |
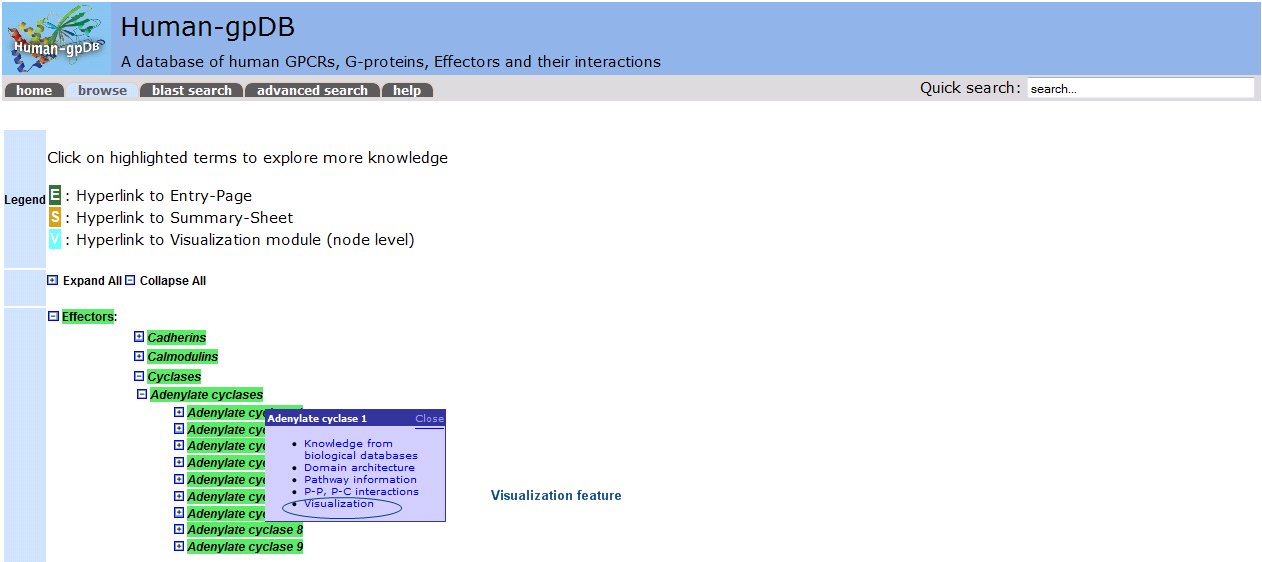
By left clicking we generate a popup window that refers to the level of the tree. This popup window provides us with the following options:
- Knowledge from biological databases: lists the proteins of the level and gives a summary for each protein.
- Domain architecture: gives information about the domain architecture for each member of the level using information from SMART.
- Pathway information: gives information about the pathways in which the members of this level participate based on information from KEGG.
- P-P, P-C interactions: shows a network of all the known P-P and P-C interactions using STITCH.
- Visualization: The visualization module in Human-gpDB was designed in a such a way that to give maximum flexibility to the user to visualize the interactions between different knowledge domains (GPCRs, G-proteins, Effectors, drugs related to GPCRs, drugs related to G-proteins, drugs related to Effectors) at different levels (depths) by taking the advantage of hierarchical categories of GPCRs, G-proteins, Effectors. Two visualization tools were provided, Medusa for 2D representation and Arena3D for 3D representation of the network. In the case of Arena3D, we provide a downloadable inputfile for Arena3D tool, so the user must download and install locally Arena3D and then use the given input file to view 3D representation of the interactions.

The text search allows the user to search the bioentities by auto completing forms by using the name, the IDs, the gene names and the cross-references provided by our database.
|
|
|
 5. What is the BLAST option and how do I use it? 5. What is the BLAST option and how do I use it? |
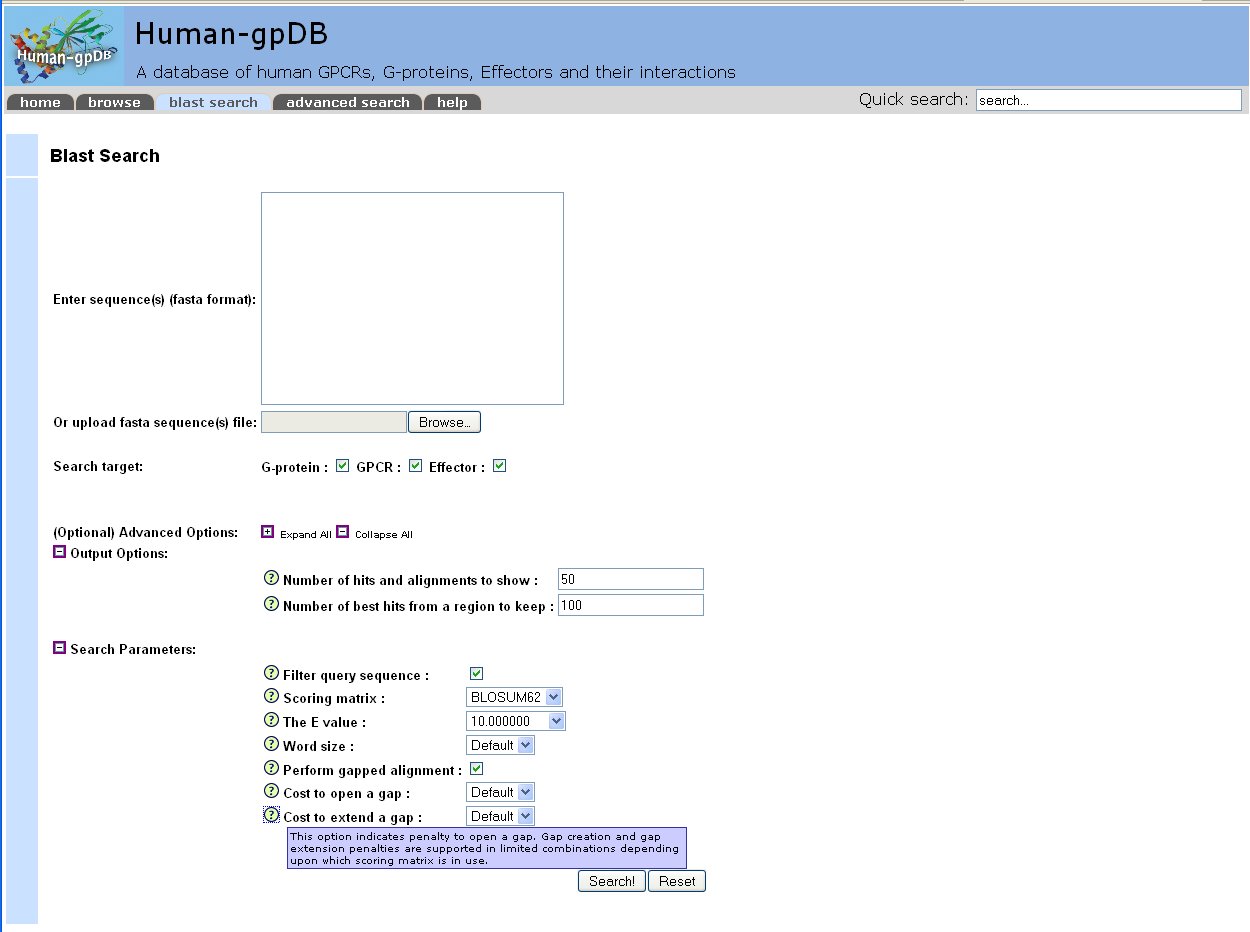
With the BLAST search tool, the user may submit a sequence and search the database for finding homologues. The user has the option to choose whether to perform the BLAST search against GPCRs sequences, G-proteins sequences and/or Effectors sequences. The input for the BLAST application is the sequence in standard FASTA format.
The output of the BLAST query consists of a list of sequences in the database having significant E-values in a local pair wise alignment, ranked by statistical significance. In the output, are also listed the range of residues in which the alignment occurs, in both the target and the query sequence, the number of identical and similar residues in the alignment and the E-value of the alignment.
|
|
|
 6. What is Advanced Search and how do I use it? 6. What is Advanced Search and how do I use it? |
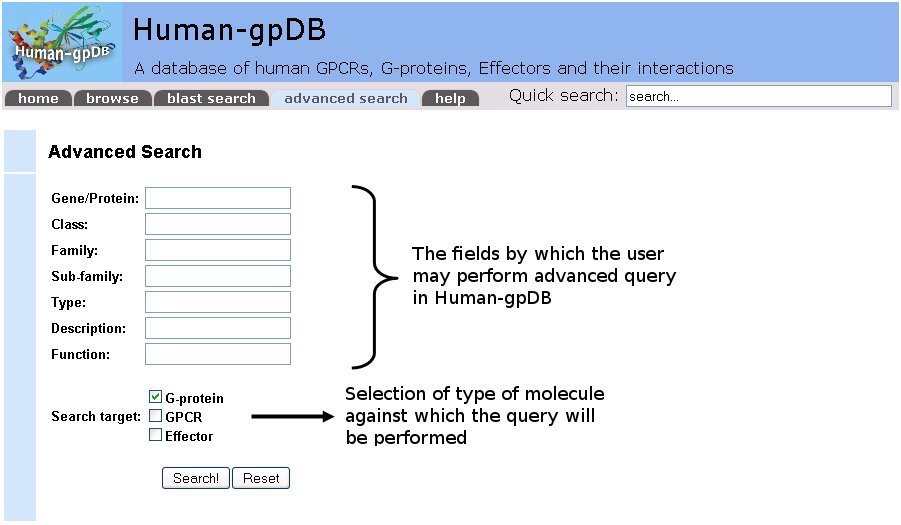
This feature gives users the option to search the given fields in the database. The user can enter any word in one or more of the available boxes under the name: Gene/Protein, Class, Family, Subfamily, Type, Description and Function. Expressions in separate search fields are combined with the AND operator, so every entry of the result set will satisfy the expressions of all the search fields the user has chosen. The user has the option to choose whether the query will be performed against the GPCRs, the G-proteins or the Effectors included in the database.
|
|
|
 7. How did we get protein features and what information do we provide for the individual proteins? 7. How did we get protein features and what information do we provide for the individual proteins? |
The protein features are obtained by Dasty DAS service and most of the links provided by UNIPROT concerning each protein.
The main information we provide for individual proteins are: protein name, description, classification, gene name, sequence, cross references, interactions, orthology information.
|
|
|
 8. How does the visualization help me? 8. How does the visualization help me? |
We provide 2 different visualization tools for visualizing the bioentities and their interactions in both 2D and 3D dimensions. These are Medusa and Arena3D.
2D visualization is supported by Medusa Java applet where proteins and how they are connected with other proteins is visualized. Medusa requires Java 6 or higher version for the applet to run on the client side. It comes with various layout and clustering algorithms that are able to minimize the crossovers between the lines and make the network easier to understand.
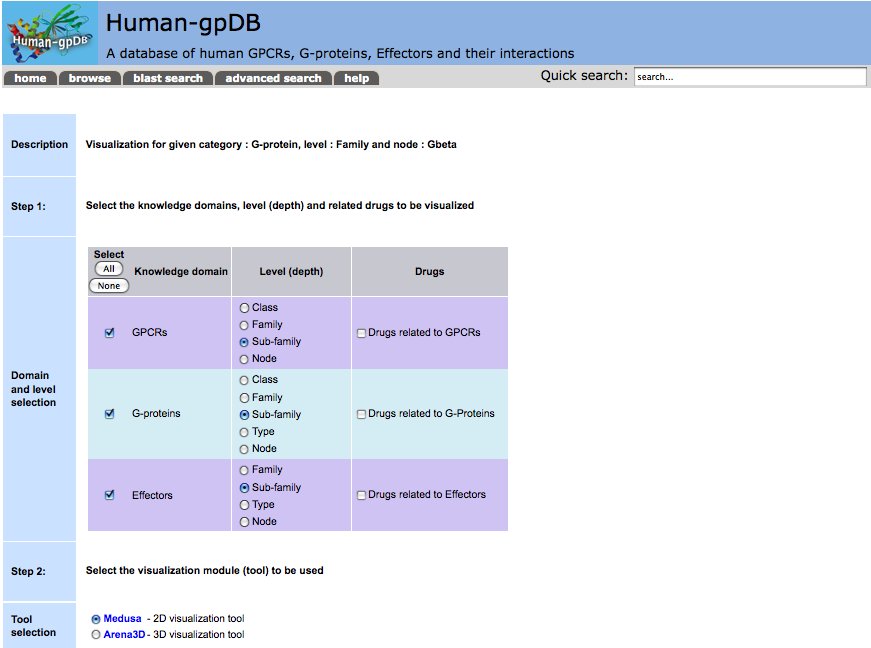
3D visualization is supported by Arena3D application. Arena3D is a standalone application so users need to download the software first to use it. Human-gpDB provides input files for Arena3D, which contain information about how proteins are connected among each other. We separate GPCRS, G-Proteins and Effectors onto different layers by using a multilayered graph approach and we try to show how they are connected in 3D space. Arena3D is a nice application to support larger networks whereas Medusa is restricted to smaller ones. Arena3D like Medusa comes with a variety of layout and clustering algorithms to make the network easier and the extraction of information easier. Arena3D requires Java 6 or higher together with Java3D 1.5.1 API to run on the clients side.
|
|
|
 9. I have some problems using Human-gpDB? Whom should I contact? 9. I have some problems using Human-gpDB? Whom should I contact? |
Human-gpDB is a collaborative project between the following two groups:
1. Prof. Stavros J. Hamodrakas, Department of Cell Biology and Biophysics, University of Athens, Greece (http://biophysics.biol.uoa.gr).
2. Dr. Reinhard Schneider, Structural and Computational Biology, EMBL, Heidelberg, Germany (http://www.embl.de/research/units/scb/schneider_reinhard/index.html).
The scientific supervisors were Professor Stavros J. Hamodrakas together with Dr. Reinhard Schneider and Assistant Professor Pantelis Bagos, formerly at the University of Athens, now at the University of Central Greece, Lamia, Greece. The data was collected by Margarita Theodoropoulou (who also helped in the web application and database development) and Christos Stabolakis. Venkata P. Satagopam was the main developer behind the web application and data integration. Dr. Georgios Pavlopoulos worked on the Visualization tools and Dr. Nikolaos Papandreou helped in the development of the application.
E-mails:
venkata.satagopam@uni.lu
mtheodo@biol.uoa.gr
pavlopou@embl.de
npapand@biol.uoa.gr
pbagos@ucg.gr
reinhard.schneider@uni.lu
shamodr@biol.uoa.gr
Adresses:
Structural and Computational Biology
EMBL
Meyerhofstr. 1
19117 Heidelberg
Germany
tel: +49-6221-387528
fax: +49-6221-387517
Department of Cell Biology & Biophysics
Faculty of Biology
University of Athens
Panepistimiopolis
Athens
GR-15701 GREECE
tel: +30-210-7274931
fax: +30-210-7274254
|
|